The prediction of protein-protein interactions (PPIs) is crucial for understanding biological functions and diseases. We propose ProLLM, a novel framework that employs an LLM tailored for PPI for the first time. Specifically, we propose Protein Chain of Thought (ProCoT), which simulates the biological mechanism of signaling pathways as natural language prompts, predicting interactions between proteins through reasoning. ProLLM significantly improves prediction accuracy and generalizability compared to existing methods.
Our framework, ProLLM, employs a Protein Chain of Thought (ProCoT) approach to simulate protein signaling pathways as natural language prompts. It incorporates embedding replacement with ProtTrans vectors and instruction fine-tuning on protein knowledge datasets, leading to a deeper understanding of complex biological problems.
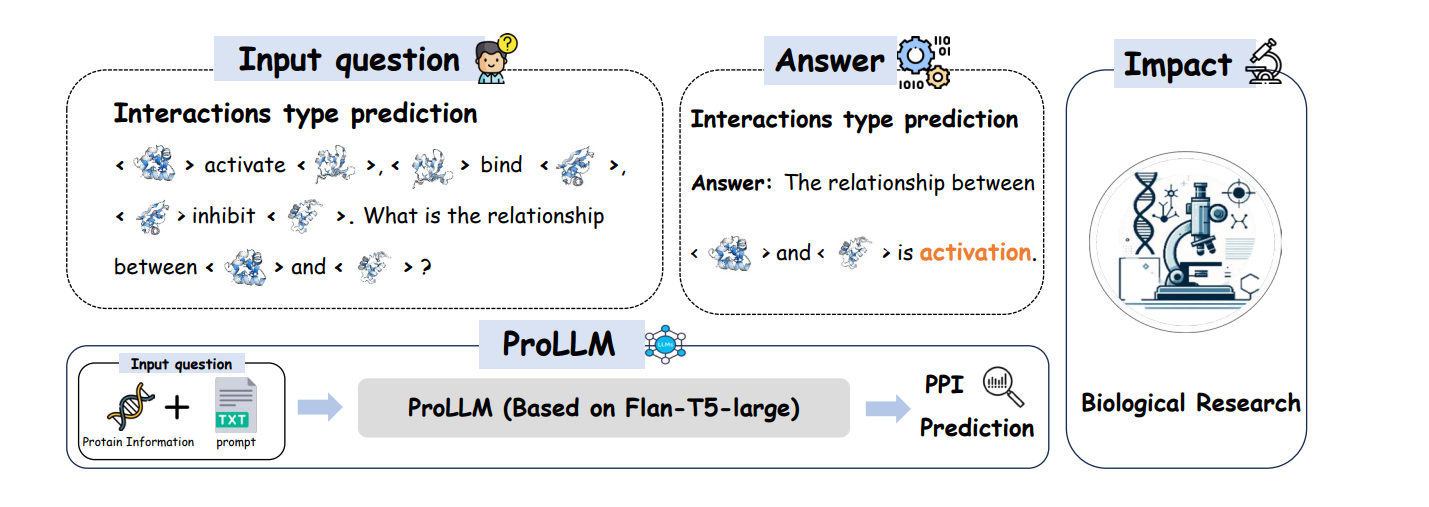
Previous approaches like CNNs and GNNs focus on direct physical interactions but fail to capture non-physical connections. LLMs offer a new avenue by representing protein-protein interactions as natural language problems. We build on methods such as ProtBERT and ProteinLM, enhancing the model's capability to reason about protein chains through ProCoT.
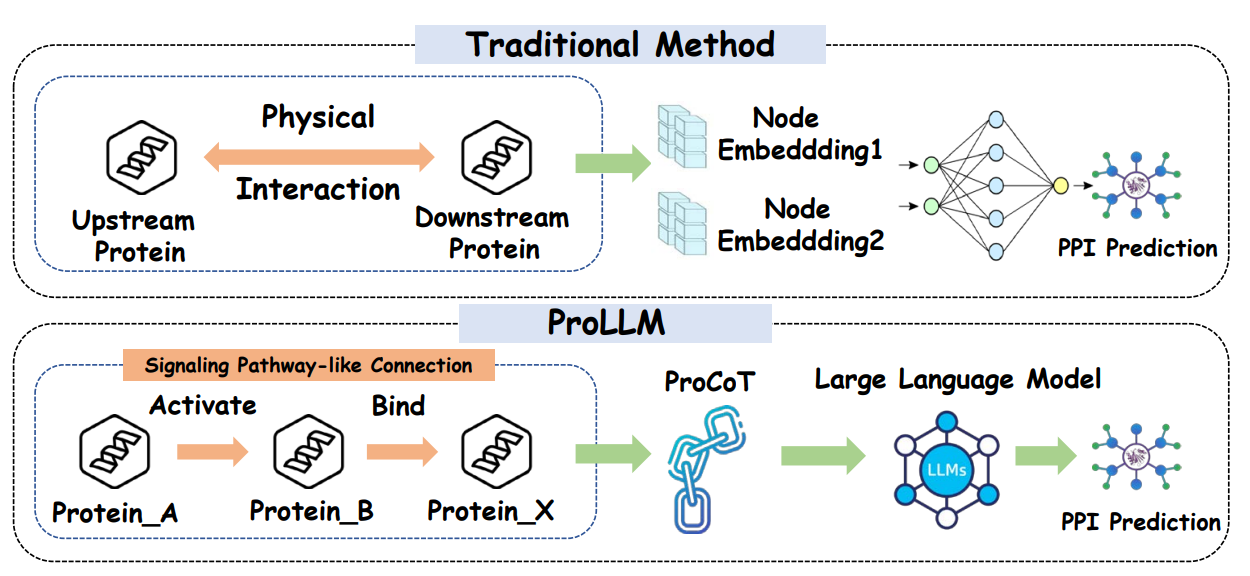
The following sections describe the detailed methodology employed in ProLLM for protein-protein interaction prediction. This framework is divided into four key sectors, each contributing to the overall performance improvement in predicting protein interactions.
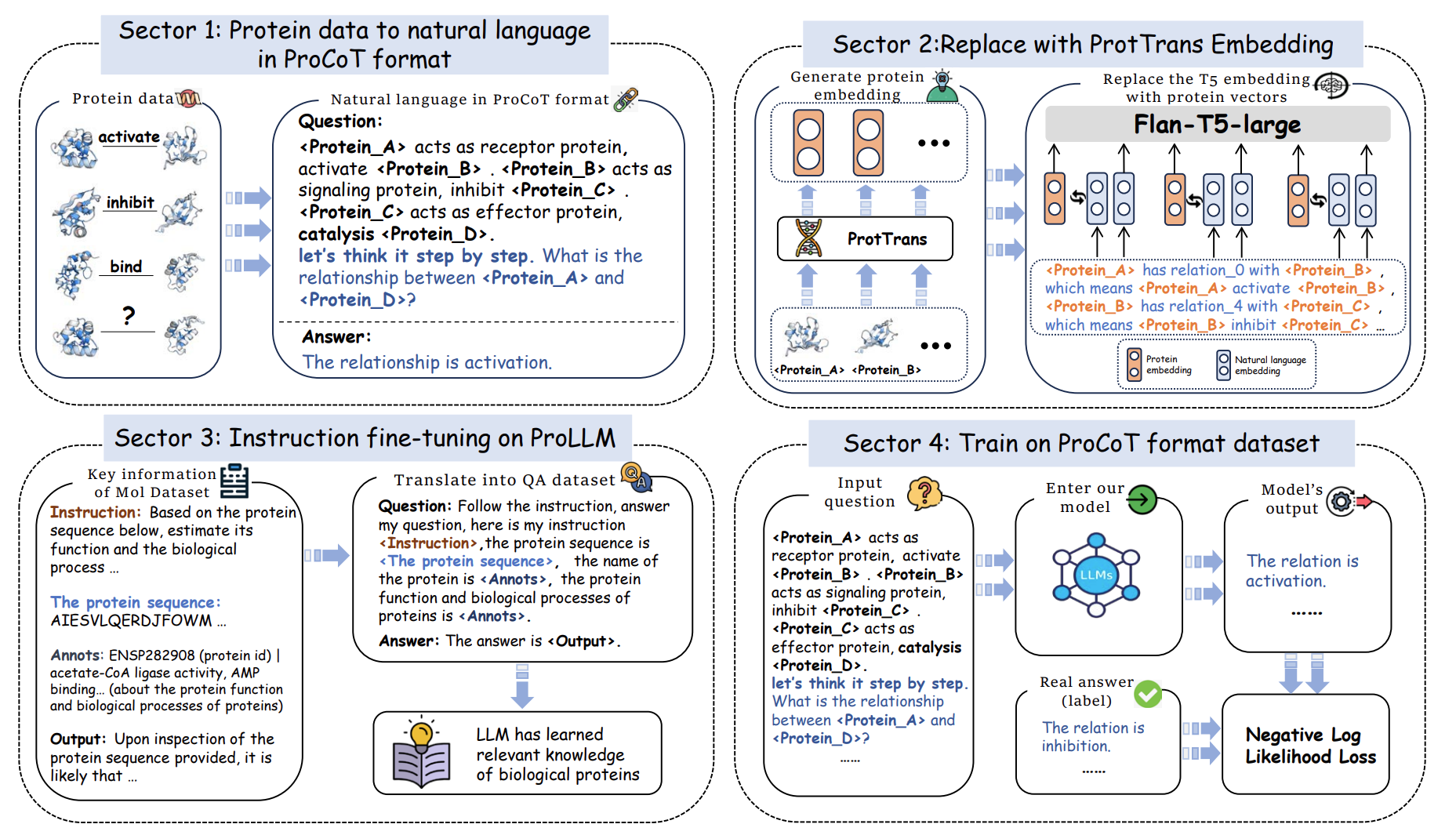
In the first step of our methodology, we transform raw protein data into natural language prompts in the Protein Chain of Thought (ProCoT) format. This process simulates the biological signaling pathways between proteins, using descriptions of actions such as "activate", "inhibit", and "bind" to represent the interactions. By converting these structured data into a natural language format, we enable the large language model (LLM) to process and reason about the relationships between proteins step by step.
For example, a typical ProCoT prompt might be: "<Protein_A> acts as a receptor protein, activate <Protein_B>. <Protein_B> acts as a signaling protein, inhibit <Protein_C>. <Protein_C> acts as an effector protein, catalysis <Protein_D>. What is the relationship between <Protein_A> and <Protein_D>?" This helps the model simulate the biological reasoning chain, leading to more accurate interaction predictions.
In this step, we enhance the LLM’s understanding of protein sequences by replacing standard language embeddings with protein-specific embeddings generated from the ProtTrans model. ProtTrans, a large-scale pre-trained model, generates embeddings that capture the biophysical and structural properties of proteins.
By integrating ProtTrans embeddings directly into the LLM, we provide it with richer biological information, allowing it to better understand the relationships between proteins. This step is crucial for improving the model’s performance in recognizing complex protein interactions beyond simple textual descriptions.
After embedding replacement, we fine-tune the ProLLM on a protein knowledge dataset using an instruction-based approach. This fine-tuning process allows the model to gain specialized knowledge about proteins, including their functions and biological processes. We use instruction datasets like Mol-Instructions, which provide detailed prompts and corresponding answers about protein functions.
For instance, a fine-tuning instruction might ask: "Based on the following protein sequence, predict its function." The model is then able to infer important details about the protein’s role in various biological processes, further enhancing its predictive power when applied to new protein-protein interaction tasks.
In the final step, we train the ProLLM on datasets formatted in the ProCoT structure. The dataset contains complex signaling pathways between proteins, where the model learns to predict the type of interaction (e.g., activation, inhibition) between two proteins.
During training, the model is presented with input questions such as: "
We conducted extensive experiments to evaluate the performance of ProLLM compared to existing models. The comparison includes models based on Convolutional Neural Networks (CNNs), Graph Neural Networks (GNNs), and other Large Language Models (LLMs). The results were evaluated on several widely used Protein-Protein Interaction (PPI) datasets, including Human, SHS27K, SHS148K, and STRING.
ProLLM outperforms existing models in terms of both accuracy and generalizability. The key advantage of ProLLM lies in its ability to model complex protein signaling pathways through the ProCoT format, which allows it to capture indirect interactions between proteins more effectively than traditional models.
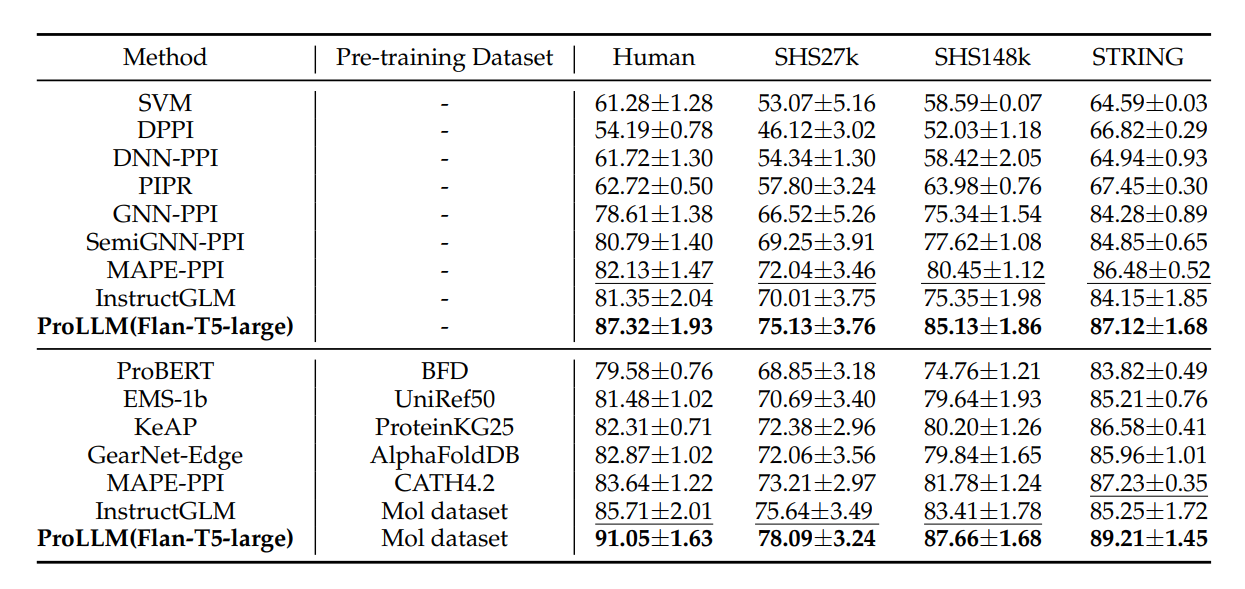
The comparison results show that ProLLM achieves significantly higher micro-F1 scores across all datasets. For example, on the Human dataset, ProLLM achieved a micro-F1 score of 91.05, surpassing GNN-PPI's score of 78.61 and CNN-based methods like DPPI's score of 54.19. Similar improvements were observed in SHS27K, SHS148K, and STRING datasets, where ProLLM consistently demonstrated better performance due to its ability to model non-physical connections between proteins.
The following table and figure summarize the comparative performance of different models, highlighting ProLLM's superiority in both pre-trained and non-pre-trained setups.
To better understand the contribution of each component in ProLLM, we conducted an ablation study. The ablation experiments were performed by systematically removing or altering key components of the model to assess their impact on overall performance. The components studied include:
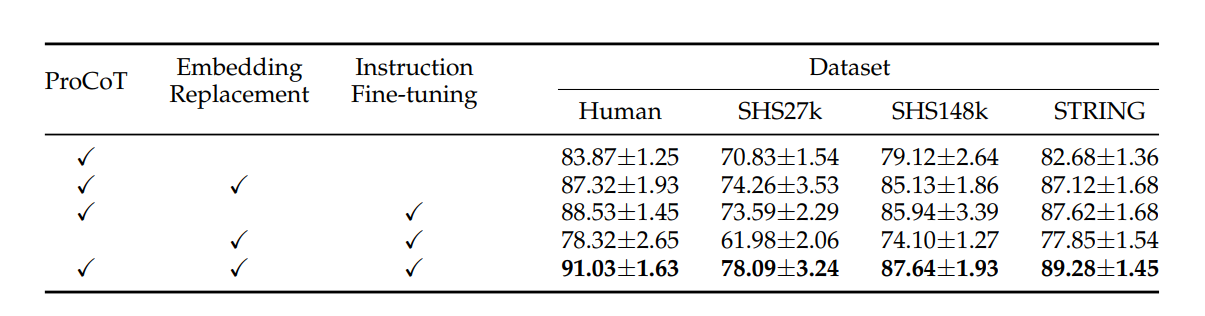
The results of the ablation study, as shown in the figure, demonstrate the importance of each component in ProLLM's overall performance. Removing the ProCoT format resulted in the largest decrease in performance, as the model was no longer able to reason about signaling pathways. Without ProtTrans embeddings, the model’s understanding of protein sequences was significantly reduced, resulting in lower micro-F1 scores across all datasets. Lastly, the removal of instruction fine-tuning also caused a notable drop in performance, particularly in tasks that required understanding protein functions.
These findings highlight the critical role each component plays in enhancing the predictive capabilities of ProLLM, particularly the ProCoT format and embedding replacement with ProtTrans.
In conclusion, ProLLM is a powerful framework for predicting protein-protein interactions. It leverages the power of LLMs to represent signaling pathways in a natural language format, significantly enhancing accuracy and generalizability.
We would like to express our gratitude to all those who contributed to this project. Special thanks to Dr. Wenyue Hua, Dr. Kai Mei, and Dr. Taowen Wang for their insightful discussions and suggestions during the development of this work. We also acknowledge the support from our institutions, including Rutgers University, the University of Liverpool, Peking University, MIT, and the New Jersey Institute of Technology, for providing the necessary resources and computational infrastructure.
@article{jin2024prollm,
title={ProLLM: Protein Chain-of-Thoughts Enhanced LLM for Protein-Protein Interaction Prediction},
author={Jin, Mingyu and Xue, Haochen and Wang, Zhenting and Kang, Boming and Ye, Ruosong and Zhou, Kaixiong and Du, Mengnan and Zhang, Yongfeng},
journal={bioRxiv},
pages={2024--04},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}